Volume 34, Issue 8 (11-2023)
Studies in Medical Sciences 2023, 34(8): 479-485 |
Back to browse issues page
Research code: 4010602
Ethics code: IR.KUMS.REC.1401.405
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Foroughi A, Ghadiri K, Bozorgomid A, Chegeneh Lorestani R, Jafari S. An Investigation on the Prevalence of Escherichia Albertii in Urine and Stool Samples of Patients in Kermanshah, Iran. Studies in Medical Sciences 2023; 34 (8) :479-485
URL: http://umj.umsu.ac.ir/article-1-6054-en.html
URL: http://umj.umsu.ac.ir/article-1-6054-en.html
Azadeh Foroughi * 

 , Keyghobad Ghadiri
, Keyghobad Ghadiri 

 , Arezoo Bozorgomid
, Arezoo Bozorgomid 

 , Roya Chegeneh Lorestani
, Roya Chegeneh Lorestani 

 , Somayeh Jafari
, Somayeh Jafari 




 , Keyghobad Ghadiri
, Keyghobad Ghadiri 

 , Arezoo Bozorgomid
, Arezoo Bozorgomid 

 , Roya Chegeneh Lorestani
, Roya Chegeneh Lorestani 

 , Somayeh Jafari
, Somayeh Jafari 


Department of Pathobiology & Basic Science, Veterinary Science Faculty, Razi University, Kermanshah, Iran (Corresponding Author) , a.foroughi@razi.ac.ir
Full-Text [PDF 291 kb]
(769 Downloads)
| Abstract (HTML) (2669 Views)
Table 1. The reaction components for PCR
Table 2. Results of culture and PCR tests for detection of E. albertii in samples
Full-Text: (397 Views)
Introduction
Diarrheal diseases are known as of the health problems around the world, especially in developing countries, which are of the main causes of death in developing countries and the second cause of death for children worldwide. They were responsible for 1.1 million deaths of children aged 5 and above and 1.5 million deaths of children under 5 years of age in 2009 (1, 2). The most important cause of diarrhea is Escherichia coli. This bacterium is also the main cause of Urinary Tract Infections (UTIs) in humans, especially women (3). On the other hand, studies have shown that Urinary Tract Infections are related to fertility disorders in men, among which Escherichia coli has been more and more noticed as the most common microorganism of Urinary Tract Infections (4, 5).
Until recently, there were six species in the Escherichia genus, named E. coli, E. fergusonii, E. blattae, E. adecarboxylata, E. hermanii and E. vulneris, among which, Escherichia coli is the most important (6, 7). Recently, another species named E. albertii has been introduced by Huys et al. in relation to diarrheal diseases in Bangladeshi children (8). At first, the bacterium was diagnosed as atypical H. alvei from stool samples of children under 5 years old with diarrhea in 1990 (8, 9). Based on DNA-DNA hybridization analysis, Hafnia alvei-like strains were classified as a new species as Escherichia albertii (9).
E. albertii has been isolated from the urine and stool of humans, pigs, and bats. It has also been isolated from 14 bird species belonging to 10 families (10, 11). Due to the high prevalence of E. albertii among chickens and since the bacterium can rarely be isolated from the stool of asymptomatic human, chickens are considered as a potential source of contamination for humans (12, 13). In the United States, E. albertii has been proposed as a possible etiological agent in foodborne diseases (14, 15). The results of a study done along 2003-2014 showed that out of a total of 282 E. albertii isolated, more than half of them (144 cases) have been associated with diarrhea or gastroenteritis in humans (16). The results of recent studies proved the existence of a strong relationship between E. albertii and disease outbreaks (17-20).
The exact clinical significance and prevalence of Escherichia albertii are unknown to some extent. The reason is often attributed to the impossibility of identifying this species using conventional biochemical identification test, which may be mistakenly diagnosed as Shigella, Escherichia coli, or Hafnia alvei because of its genetic and phenotypic similarity to these pathogens. For instance, E. albertii commonly carries the eae gene that encodes intimin, an important virulence factor also harbored by pathogenic subgroups of E. coli (9, 21, 22). About biochemical tests, E. albertii strains show variable properties. Because of this, to date, there is no standard method to isolation the bacterium (23). E. albertii is not mentioned in databases of API and Vitek (24).
TIt can be said that in order to know more about the characteristics of E. albertii as a human enteric pathogen and to know its prevalence, more analyzes of different sources, different hosts and more accurate identification methods are needed (18). Therefore, the aim of the present study was to investigate the prevalence of E. albertii in the urine and stool samples of UTI patients presenting in Kermanshah hospitals using culture and conventional PCR. As far as the authors know, this is the first study in Iran to detect E. albertii by using specific primer for cdt gene.
Materials & Methods
Study design:
This cross-sectional study was conducted to isolate and identify Escherichia albertii by culture and PCR methods from urine and stool samples of UTI patients in Kermanshah City. All study procedures were approved by the Ethics Committee of the Kermanshah University of Medical Sciences, Iran (IR.KUMS.REC.1401.405). In a period of 2 months (Summer 2022), 400 positive samples (200 urine and 200 stool samples) for Enterobacteriaceae (Gram-negative, non-spore-forming rods, glucose-fermentor, oxidase-negative and nitrate-reducing bacteria) were collected from the patients referred to hospitals in Kermanshah City (Mainly Imam Reza Hospital). The samples were transferred to the microbiology laboratory of Faculty of Veterinary Medicine, Razi University for additional culture and confirmation.
Bacterial cultures:
Cultivation on blood agar and EMB agar media (Quelab, Canada) was done. Routine differential tests including TSI, SIM, MR-VP and Simon citrate (Quelab, Canada) were carried out according to the method provided by Foroughi et al. (25). After that, the samples were enriched in peptone buffer (BPW- Ibresco, Iran) for 24 hours and then cultured on XRM-MC medium consisted of McConkey agar (Ibresco, Iran) with rhamnose, xylose and melibiose sugars at 1% each (Merck, Germany) and after 24 hours, white colonies (Suspicious for E. albertii) were selected and stored at -20°C for further PCR test according to the method provided by Hinenoya et al. (26).
Genomic DNA Extraction and PCR Assay:
DNA extraction was done by boiling method. Then, the isolates were evaluated for the presence of Eacdt gene (Encoding a cytolethal distending toxin in E. albertii). The PCR was used to amplify the gene using specific primers (5’→3’): Eacdt-F: GCTTAACTGGATGATTCTTG and Eacdt -R: CTATTTCCCATCCAATAGTCT (24). Primers and all the other PCR reagents were prepared from CinnaGen company (Iran). The total PCR reaction volume was 20 μl. The reaction components and their volumes are shown in the table 1.
Diarrheal diseases are known as of the health problems around the world, especially in developing countries, which are of the main causes of death in developing countries and the second cause of death for children worldwide. They were responsible for 1.1 million deaths of children aged 5 and above and 1.5 million deaths of children under 5 years of age in 2009 (1, 2). The most important cause of diarrhea is Escherichia coli. This bacterium is also the main cause of Urinary Tract Infections (UTIs) in humans, especially women (3). On the other hand, studies have shown that Urinary Tract Infections are related to fertility disorders in men, among which Escherichia coli has been more and more noticed as the most common microorganism of Urinary Tract Infections (4, 5).
Until recently, there were six species in the Escherichia genus, named E. coli, E. fergusonii, E. blattae, E. adecarboxylata, E. hermanii and E. vulneris, among which, Escherichia coli is the most important (6, 7). Recently, another species named E. albertii has been introduced by Huys et al. in relation to diarrheal diseases in Bangladeshi children (8). At first, the bacterium was diagnosed as atypical H. alvei from stool samples of children under 5 years old with diarrhea in 1990 (8, 9). Based on DNA-DNA hybridization analysis, Hafnia alvei-like strains were classified as a new species as Escherichia albertii (9).
E. albertii has been isolated from the urine and stool of humans, pigs, and bats. It has also been isolated from 14 bird species belonging to 10 families (10, 11). Due to the high prevalence of E. albertii among chickens and since the bacterium can rarely be isolated from the stool of asymptomatic human, chickens are considered as a potential source of contamination for humans (12, 13). In the United States, E. albertii has been proposed as a possible etiological agent in foodborne diseases (14, 15). The results of a study done along 2003-2014 showed that out of a total of 282 E. albertii isolated, more than half of them (144 cases) have been associated with diarrhea or gastroenteritis in humans (16). The results of recent studies proved the existence of a strong relationship between E. albertii and disease outbreaks (17-20).
The exact clinical significance and prevalence of Escherichia albertii are unknown to some extent. The reason is often attributed to the impossibility of identifying this species using conventional biochemical identification test, which may be mistakenly diagnosed as Shigella, Escherichia coli, or Hafnia alvei because of its genetic and phenotypic similarity to these pathogens. For instance, E. albertii commonly carries the eae gene that encodes intimin, an important virulence factor also harbored by pathogenic subgroups of E. coli (9, 21, 22). About biochemical tests, E. albertii strains show variable properties. Because of this, to date, there is no standard method to isolation the bacterium (23). E. albertii is not mentioned in databases of API and Vitek (24).
TIt can be said that in order to know more about the characteristics of E. albertii as a human enteric pathogen and to know its prevalence, more analyzes of different sources, different hosts and more accurate identification methods are needed (18). Therefore, the aim of the present study was to investigate the prevalence of E. albertii in the urine and stool samples of UTI patients presenting in Kermanshah hospitals using culture and conventional PCR. As far as the authors know, this is the first study in Iran to detect E. albertii by using specific primer for cdt gene.
Materials & Methods
Study design:
This cross-sectional study was conducted to isolate and identify Escherichia albertii by culture and PCR methods from urine and stool samples of UTI patients in Kermanshah City. All study procedures were approved by the Ethics Committee of the Kermanshah University of Medical Sciences, Iran (IR.KUMS.REC.1401.405). In a period of 2 months (Summer 2022), 400 positive samples (200 urine and 200 stool samples) for Enterobacteriaceae (Gram-negative, non-spore-forming rods, glucose-fermentor, oxidase-negative and nitrate-reducing bacteria) were collected from the patients referred to hospitals in Kermanshah City (Mainly Imam Reza Hospital). The samples were transferred to the microbiology laboratory of Faculty of Veterinary Medicine, Razi University for additional culture and confirmation.
Bacterial cultures:
Cultivation on blood agar and EMB agar media (Quelab, Canada) was done. Routine differential tests including TSI, SIM, MR-VP and Simon citrate (Quelab, Canada) were carried out according to the method provided by Foroughi et al. (25). After that, the samples were enriched in peptone buffer (BPW- Ibresco, Iran) for 24 hours and then cultured on XRM-MC medium consisted of McConkey agar (Ibresco, Iran) with rhamnose, xylose and melibiose sugars at 1% each (Merck, Germany) and after 24 hours, white colonies (Suspicious for E. albertii) were selected and stored at -20°C for further PCR test according to the method provided by Hinenoya et al. (26).
Genomic DNA Extraction and PCR Assay:
DNA extraction was done by boiling method. Then, the isolates were evaluated for the presence of Eacdt gene (Encoding a cytolethal distending toxin in E. albertii). The PCR was used to amplify the gene using specific primers (5’→3’): Eacdt-F: GCTTAACTGGATGATTCTTG and Eacdt -R: CTATTTCCCATCCAATAGTCT (24). Primers and all the other PCR reagents were prepared from CinnaGen company (Iran). The total PCR reaction volume was 20 μl. The reaction components and their volumes are shown in the table 1.
Table 1. The reaction components for PCR
| Components | Description | Volume (µl) |
| Primers (10 µM Solution) |
Eacdt-F Eacdt -R |
1 1 |
| Master Mix | Premixed | 16 |
| Template | DNA | 2 |
| Final volume | 20 |
The reaction was performed by initial denaturation for 4 min at 95°C, then 40 cycles of denaturation at 94°C for 30 seconds, annealing at 50°C for 30 sec, and extension at 72°C for 30 seconds (26) in the thermal cycler (Padide-Nogen-Pars, Iran). The PCR products were kept at 4°C until the electrophoresis was run.
Gel Electrophoresis:
PCR products (5 µl) were subjected to electrophoresis. Electrophoresis was run on a 2% agarose gel prepared with TBE buffer 0.5% and 10 µl ethidium bromide (Sinaclon, Iran). A constant voltage of 90 V for 1.5 hours and the agarose gels were imaged under UV light (316 nm) using the transilluminator (Fargene, Iran) and analyzed for band sizes. The expected size of the PCR product was 449 bp.
Results
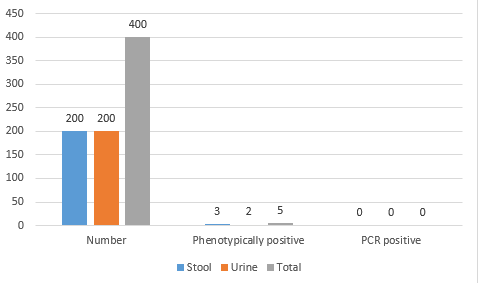
Out of four hundred samples (200 urine and 200 stool samples) positive for Enterobacteriaceae, only 5 samples (1.25%) were presumptive positive in the final culture on XRM-MacConkey agar (Suspicious for E. albertii). Three samples (0.75%) were from stool and 2 samples (0.5%). None of them were then confirmed as E. albertii by the PCR (Table 2 and Figure 1). These 5 samples were diagnosed as E. coli by performing biochemical tests, but PCR was not done to confirm it.
Gel Electrophoresis:
PCR products (5 µl) were subjected to electrophoresis. Electrophoresis was run on a 2% agarose gel prepared with TBE buffer 0.5% and 10 µl ethidium bromide (Sinaclon, Iran). A constant voltage of 90 V for 1.5 hours and the agarose gels were imaged under UV light (316 nm) using the transilluminator (Fargene, Iran) and analyzed for band sizes. The expected size of the PCR product was 449 bp.
Results
Out of four hundred samples (200 urine and 200 stool samples) positive for Enterobacteriaceae, only 5 samples (1.25%) were presumptive positive in the final culture on XRM-MacConkey agar (Suspicious for E. albertii). Three samples (0.75%) were from stool and 2 samples (0.5%). None of them were then confirmed as E. albertii by the PCR (Table 2 and Figure 1). These 5 samples were diagnosed as E. coli by performing biochemical tests, but PCR was not done to confirm it.
Table 2. Results of culture and PCR tests for detection of E. albertii in samples
| Samples | Number | Phenotypically positive (Number/Percentage) |
PCR positive (Number/Percentage) |
| Stool | 200 | 3 (0.75%) | 0 (0%) |
| Urine | 200 | 2 (0.5%) | 0 (0%) |
| Total | 400 | 5 (1.25%) | 0 (0%) |

Fig. 1. Comparison of presumptive positive E. albertii in urine and stool samples.
Discussion
Despite limited studies on E. albertii in Iran, many efforts have been made to detect and isolate the bacterium from various human, animal, poultry and food samples around the world. It is important to detect this bacterium in samples and determine its prevalence because its antibiotic resistance and virulence factors may be different from other similar bacteria, which shows its importance in the clinical treatment and epidemiology of the diseases caused by.
A new study showed that out of 296 stool samples from children with gastroenteritis, 11.8% were identified as E. albertii in culture, all of which were confirmed by PCR method using eae, lysP, and mdh primers. The highest prevalence was in infants aged 0 to 6 months (24). Also, Sulaiman et al. (2021) reported the prevalence of the bacterium as 1.1 to 1.3% in patients with gastroenteritis and 0% in apparently healthy individuals. In addition, the prevalence was declared equal between men and women and mostly in children aged 0 to 10 years (27). Zaki et al. (2021) investigated 100 phenotypically confirmed isolates as E. coli from children with UTI and proved 7 samples as E. albertii by the molecular technique. In this study, primers uidA (to confirm E. coli) and mdh-lysP (To confirm E. albertii) were used to examine (28). In Iran, a similar study was conducted in Kermanshah in 2018 on the urine and stool samples of patients, during which, out of 100 phenotypically confirmed samples as E. coli, 6 samples were confirmed as E. albertii and 94 samples were confirmed as E. coli by the PCR using the same primers as Zaki’s (25).
In the present study, out of 400 samples of urine and stool culture that were positive for Enterobacteriaceae; finally 5 samples were suspected to be E. albertii by the biochemical tests described by Hinenoya et al. (2019). None of them was confirmed by PCR test using the primer introduced by him. The reason for not detecting this bacterium in the present study may be due to the different types of primers used. Specific primers for mdh and lysP genes have been used in many studies and resulted in bacterial detection (15, 17, 20, 29). Hinenoya et al. (2019) introduced a primer to detect the E. albertii cytolethal distending toxin (Eacdt) gene for the first time with 100% sensitivity and specificity for E. albertii detection (26). In a study done by Muchaamba et al. in 2022, 319 samples were examined by this primer as in-silico PCR method in which 310 samples were confirmed as E. albertii (96.1%). The reason for the difference between these two studies is attributed to the limited samples (64 suspected E. albertii isolates) examined in the study done by Hinenoya et al. (23).
Conclusion
As far as the authors know, no other study has been carried out with this primer for the molecular detection of E. albertii. It seems that the use of previously introduced primers (specific for mdh and lysP genes) is more efficient for the detection of this bacterium. However, further studies are recommended to confirm it. On the other hand, less sensitive primers could lead to underestimation of the prevalence of E. albertii in Iran and elsewhere. One of the limitations of the study is that the study is cross-sectional and conducted in a geographically limited area, which can limit the overall estimate of the prevalence of this bacterium. Of other limitations are small sample size and the use of a single PCR primer. So, more researches are needed to investigate this.
Acknowledgement
The authors would like to express their gratitude to Mrs. Nikpey, the respected expert of the microbiology laboratory of Faculty of Veterinary Medicine, Razi University, for her sincere cooperation.
Source of funding
This project has no financial sponsor and its costs have been paid personally.
Conflict of interest
The authors declare that there is no conflict of interest.
Ethical consideration
All study procedures were approved by the Ethics Committee of the Kermanshah University of Medical Sciences, Iran (IR.KUMS.REC.1401.405).
Data availability
The datasets during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Despite limited studies on E. albertii in Iran, many efforts have been made to detect and isolate the bacterium from various human, animal, poultry and food samples around the world. It is important to detect this bacterium in samples and determine its prevalence because its antibiotic resistance and virulence factors may be different from other similar bacteria, which shows its importance in the clinical treatment and epidemiology of the diseases caused by.
A new study showed that out of 296 stool samples from children with gastroenteritis, 11.8% were identified as E. albertii in culture, all of which were confirmed by PCR method using eae, lysP, and mdh primers. The highest prevalence was in infants aged 0 to 6 months (24). Also, Sulaiman et al. (2021) reported the prevalence of the bacterium as 1.1 to 1.3% in patients with gastroenteritis and 0% in apparently healthy individuals. In addition, the prevalence was declared equal between men and women and mostly in children aged 0 to 10 years (27). Zaki et al. (2021) investigated 100 phenotypically confirmed isolates as E. coli from children with UTI and proved 7 samples as E. albertii by the molecular technique. In this study, primers uidA (to confirm E. coli) and mdh-lysP (To confirm E. albertii) were used to examine (28). In Iran, a similar study was conducted in Kermanshah in 2018 on the urine and stool samples of patients, during which, out of 100 phenotypically confirmed samples as E. coli, 6 samples were confirmed as E. albertii and 94 samples were confirmed as E. coli by the PCR using the same primers as Zaki’s (25).
In the present study, out of 400 samples of urine and stool culture that were positive for Enterobacteriaceae; finally 5 samples were suspected to be E. albertii by the biochemical tests described by Hinenoya et al. (2019). None of them was confirmed by PCR test using the primer introduced by him. The reason for not detecting this bacterium in the present study may be due to the different types of primers used. Specific primers for mdh and lysP genes have been used in many studies and resulted in bacterial detection (15, 17, 20, 29). Hinenoya et al. (2019) introduced a primer to detect the E. albertii cytolethal distending toxin (Eacdt) gene for the first time with 100% sensitivity and specificity for E. albertii detection (26). In a study done by Muchaamba et al. in 2022, 319 samples were examined by this primer as in-silico PCR method in which 310 samples were confirmed as E. albertii (96.1%). The reason for the difference between these two studies is attributed to the limited samples (64 suspected E. albertii isolates) examined in the study done by Hinenoya et al. (23).
Conclusion
As far as the authors know, no other study has been carried out with this primer for the molecular detection of E. albertii. It seems that the use of previously introduced primers (specific for mdh and lysP genes) is more efficient for the detection of this bacterium. However, further studies are recommended to confirm it. On the other hand, less sensitive primers could lead to underestimation of the prevalence of E. albertii in Iran and elsewhere. One of the limitations of the study is that the study is cross-sectional and conducted in a geographically limited area, which can limit the overall estimate of the prevalence of this bacterium. Of other limitations are small sample size and the use of a single PCR primer. So, more researches are needed to investigate this.
Acknowledgement
The authors would like to express their gratitude to Mrs. Nikpey, the respected expert of the microbiology laboratory of Faculty of Veterinary Medicine, Razi University, for her sincere cooperation.
Source of funding
This project has no financial sponsor and its costs have been paid personally.
Conflict of interest
The authors declare that there is no conflict of interest.
Ethical consideration
All study procedures were approved by the Ethics Committee of the Kermanshah University of Medical Sciences, Iran (IR.KUMS.REC.1401.405).
Data availability
The datasets during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Type of Study: Research |
Subject:
میکروبیولوژی
References
1. Croxen MA, Finlay, BB. Molecular mechanisms of Escherichia coli pathogenicity. Nat Rev Microbiol 2010;8:26-38. [DOI:10.1038/nrmicro2265] [PMID]
2. Estrada-Garcia T, Lopez-Saucedo C, Thompson-Bonilla R, Abonce M, Lopez-Hernandez D, Santos JI, Rosado JL, DuPont HL, Long KZ. Association of diarrheagenic Escherichia coli Pathotypes with infection and diarrhea among Mexican children and association of atypical Enteropathogenic E. coli with acute diarrhea. J Clin Microbiol 2009;47:93-8.
https://doi.org/10.1128/JCM.01166-08 [DOI:10.1128/jcm.01166-08] [PMID] []
3. Ishii S, Meyer KP, Sadowsky MJ. Relationship between phylogenetic groups, genotypic clusters, and virulence gene profiles of Escherichia coli strains from diverse human and animal sources. Appl Environ Microbiol 2007;73:5703-10.
https://doi.org/10.1128/AEM.00275-07 [DOI:10.1128/aem.00275-07] [PMID] []
4. Abera B, Biadegelgen, F, Beyene B. Prevalence of Salmonella typhi and intestinal parasites among food handlers in Bahir Dar Town, Northwest Ethiopia. Ethiop J Health Dev 2010;24:46-50. [DOI:10.4314/ejhd.v24i1.62944]
5. Dohle GR, Colpi GM, Hargreave TB, Papp GK, Jungwirth A, Weidner WE. EAU guidelines on male infertility. Eur Urol 2005; 48:703-11. [DOI:10.1016/j.eururo.2005.06.002] [PMID]
6. Welch RA. The genus Escherichia. The Prokaryotes: Volume 6: Proteobacteria: Gamma Subclass, 2006; 60-71.
https://doi.org/10.1007/0-387-30746-X_3 [DOI:10.1007/0-387-30746-x_3]
7. Making a difference. The World Health Report 1999. Health Millions 1999;25:3-5. [PMID]
8. Huys G, Cnockaert M, Janda JM, Swings J. Escherichia albertii sp. nov., a diarrhoeagenic species isolated from stool specimens of Bangladeshi children. Escherichia albertii sp. nov., a diarrhoeagenic species isolated from stool specimens of Bangladeshi children. Int J Syst Evol Microbiol 2003;53:807-10. [DOI:10.1099/ijs.0.02475-0] [PMID]
9. Albert MJ, Alam K, Islam M, Montanaro J, Rahaman AS, Haider K, Hossain MA, Kibriya AK, Tzipori S. Hafnia alvei, a probable cause of diarrhea in humans. Infect Immun 1991;59:1507-13. [DOI:10.1128/iai.59.4.1507-1513.1991] [PMID] []
10. Perez KL, Alam MJ, Castillo A, Taylor TM. Antibiotic resistance and growth of the emergent pathogen Escherichia albertii on raw ground beef stored under refrigeration, abuse, and physiological temperature. J Food Prot 2013;76:124-8.
https://doi.org/10.4315/0362-028X.JFP-12-277 [DOI:10.4315/0362-028x.jfp-12-277] [PMID]
11. Taormina P, Taylor T, Sharma M. Growth of Escherichia albertii strains in ground turkey at three temperatures. Annual Meeting of the Institute of Food Technologists 2008. [URL]
12. Gordon DM. Reservoirs of infection: the epidemiological characteristics of an emerging pathogen, Escherichia albertii. Research School of Biology, Australian National University 2011 Apr 23. [Google Scholar]
13. Wang H, Li Q, Bai X, Xu Y, Zhao A, Sun H, et al. Prevalence of eae-positive, lactose non-fermenting Escherichia albertii from retail raw meat in China. Epidemiol Infect 2016;144:45-52.
https://doi.org/10.1017/S0950268815001120 [DOI:10.1017/s0950268815001120] [PMID] []
14. Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, et al. Food-related illness and death in the United States. Emerg Infect Dis 1999; 5:607-25. [DOI:10.3201/eid0505.990502] [PMID] []
15. Nimri LF. Escherichia albertii, a newly emerging enteric pathogen with poorly defined properties. Diagn Microbiol Infect 2013;77:91-95. [DOI:10.1016/j.diagmicrobio.2013.06.028] [PMID]
16. Grillova L, Sedláček I, Pachnikova G, Staňková E, Švec P, Holochova P, et al. Characterization of four Escherichia albertii isolates collected from animals living in Antarctica and Patagonia. J Vet Med Sci 2018; 80:138-46. [DOI:10.1292/jvms.17-0492] [PMID] []
17. Asoshima N, Matsuda M, Shigemura K, Honda M, Yoshida H, Hiwaki H, et al. Identification of Escherichia albertii as a causative agent of a foodborne outbreak occurred in 2003. Jpn J Infect Dis 2014;67:139-40. [DOI:10.7883/yoken.67.139] [PMID]
18. Ooka T, Seto K, Kawano K, Kobayashi H, Etoh Y, Ichihara S, et al. Clinical significance of Escherichia albertii. Emerg Infect Dis 2012;18:488-92. [DOI:10.3201/eid1803.111401] [PMID] []
19. Ooka T, Tokuoka E, Furukawa M, Nagamura T, Ogura Y, Arisawa K, et al. Human gastroenteritis outbreak associated with Escherichia albertii, Japan. Emerg Infect Dis 2013;19:144-6. [DOI:10.3201/eid1901.120646] [PMID] []
20. Konno T, Yatsuyanagi J, Takahashi S, Kumagai Y, Wada E, Chiba M, et al. Isolation and identification of Escherichia albertii from a patient in an outbreak of gastroenteritis. Jpn J Infect Dis 2012;65:203-7. [DOI:10.7883/yoken.65.203] [PMID]
21. Abbott SL, et al. Biochemical properties of a newly described Escherichia species, Escherichia albertii. J clin microbiol 2003;41:4852-4.
https://doi.org/10.1128/JCM.41.10.4852-4854.2003 [DOI:10.1128/jcm.41.10.4852-4854.2003] [PMID] []
22. Stock, I., et al. Natural antimicrobial susceptibility patterns and biochemical identification of Escherichia albertii and Hafnia alvei strains. Diagn Microbiol Infect Dis 2005;51:151-63. [DOI:10.1016/j.diagmicrobio.2004.10.008] [PMID]
23. Muchaamba F, Barmettler K, Treier A, Houf K, Stephan R. Microbiology and Epidemiology of Escherichia albertii-An Emerging Elusive Foodborne Pathogen. Microorganisms 2022;10:875. [DOI:10.3390/microorganisms10050875] [PMID] []
24. Ghandour AMA, Fathy R, Bakry RM, et al. Screening for Escherichia albertii in children with Gastroenteritis in Pediatric Hospital at Assiut University. Egypt J Med Microbiol 2021;30:149-55. [DOI:10.21608/ejmm.2021.201434]
25. Foroughi A, Namdari A, Rahimian-Zarif, B. Detection of Escherichia albertii in Urinary and Gastrointestinal Infections in Kermanshah, Iran. Int J Enteric Pathog 2021;9:37-42. [DOI:10.34172/ijep.2021.08]
26. Hinenoya A, Ichimura H, Yasuda N, et al. Development of a specific cytolethal distending toxin (cdt) gene (Eacdt)-based PCR assay for the detection of E. albertii. Diagn Microbiol Infect Dis 2019;95:119-24. [DOI:10.1016/j.diagmicrobio.2019.04.018] [PMID]
27. Sulaiman MA, Aminu M, Ella EE, Abdullahi IO. Prevalence and risks factors of the novel Escherichia albertii among gastroenteritis patients in Kano State, Nigeria. J Med Trop 2021;23:39-45. [DOI:10.4103/jomt.jomt_34_20]
28. Zaki ME, Eid AE, El-Kazzaz SS, El-Sabbagh AM. Molecular Study of Escherichia albertii in Pediatric Urinary Tract Infections. Open Microbiol J 2021;15:139-44. [DOI:10.2174/1874285802115010139]
29. Murakami K, Etoh Y, Tanaka E, Ichihara S, Horikawa K, Kawano K, et al. Shiga toxin 2f-producing Escherichia albertii from a symptomatic human. Jpn J Infect Dis 2014;67:204-8. [DOI:10.7883/yoken.67.204] [PMID]
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |




